输入两个字符串, ![]() =
= ![]() 和
和 ![]() =
= ![]() ,逐字输出字符串的对齐方式,这样就可以减少净惩罚 最小化 .罚款计算如下: 1.罚款
,逐字输出字符串的对齐方式,这样就可以减少净惩罚 最小化 .罚款计算如下: 1.罚款 ![]() 在字符串之间插入间隙时发生。 2.罚款
在字符串之间插入间隙时发生。 2.罚款 ![]() 由于与的字符不匹配而发生
由于与的字符不匹配而发生 ![]() 和
和 ![]() .
.
例如:
Input : X = CG, Y = CA, p_gap = 3, p_xy = 7Output : X = CG_, Y = C_A, Total penalty = 6Input : X = AGGGCT, Y = AGGCA, p_gap = 3, p_xy = 2Output : X = AGGGCT, Y = A_GGCA, Total penalty = 5Input : X = CG, Y = CA, p_gap = 3, p_xy = 5Output : X = CG, Y = CA, Total penalty = 5
关于这个问题历史的简要说明 序列比对问题是生物科学的基本问题之一,旨在寻找两个氨基酸序列的相似性。比较氨基酸对人类至关重要,因为它提供了进化和发展的重要信息。Saul B.Needeman和Christian D.Wunsch为这个问题设计了一种动态规划算法,并于1970年发表。从那时起,为了提高时间复杂度和空间复杂度,已经做了许多改进,但这些都超出了本文讨论的范围。
解决方案 我们可以用动态规划来解决这个问题。可行的解决方案是在弦中引入间隙,以便使长度相等。因为可以很容易地证明,在长度相等后增加额外的间隙只会导致惩罚的增加。
最优子结构 从最优解(例如,从给定的样本输入)可以观察到,最优解仅限于 三 候选人。 1. ![]() 和
和 ![]() . 2.
. 2. ![]() 还有缺口。 3.差距和挑战
还有缺口。 3.差距和挑战 ![]() . 最优子结构的证明。 我们很容易用矛盾来证明。允许
. 最优子结构的证明。 我们很容易用矛盾来证明。允许 ![]() 是
是 ![]() 和
和 ![]() 是
是 ![]() .假设
.假设 ![]() ,
, ![]() 有一些惩罚
有一些惩罚 ![]() ,而竞争对手将受到处罚
,而竞争对手将受到处罚 ![]() 具有
具有 ![]() . 现在,附加
. 现在,附加 ![]() 和
和 ![]() ,我们得到了与点球一致的结果
,我们得到了与点球一致的结果 ![]() .这与原始路线的最佳性相矛盾
.这与原始路线的最佳性相矛盾 ![]() . 因此,事实证明了这一点。 允许
. 因此,事实证明了这一点。 允许 ![]() 是最佳排列的惩罚吗
是最佳排列的惩罚吗 ![]() 和
和 ![]() .然后,从最优子结构,
.然后,从最优子结构, ![]() . 因此,最低处罚总额为:,
. 因此,最低处罚总额为:, ![]() .
.
重建解决方案 重建, 1.追溯已填写的表格,从 ![]() . 2.什么时候
. 2.什么时候 ![]() …..2a。如果使用案例1填充,请转到
…..2a。如果使用案例1填充,请转到 ![]() . …..2b。如果使用案例2填充,请转到
. …..2b。如果使用案例2填充,请转到 ![]() . …..2c。如果使用案例3填充,请转到
. …..2c。如果使用案例3填充,请转到 ![]() . 3.如果i=0或j=0,则将剩余子串与间隙匹配。
. 3.如果i=0或j=0,则将剩余子串与间隙匹配。
下面是上述解决方案的实现。
C++
// CPP program to implement sequence alignment // problem. #include <bits/stdc++.h> using namespace std; // function to find out the minimum penalty void getMinimumPenalty(string x, string y, int pxy, int pgap) { int i, j; // initialising variables int m = x.length(); // length of gene1 int n = y.length(); // length of gene2 // table for storing optimal substructure answers int dp[m+1][n+1] = {0}; // initialising the table for (i = 0; i <= (n+m); i++) { dp[i][0] = i * pgap; dp[0][i] = i * pgap; } // calculating the minimum penalty for (i = 1; i <= m; i++) { for (j = 1; j <= n; j++) { if (x[i - 1] == y[j - 1]) { dp[i][j] = dp[i - 1][j - 1]; } else { dp[i][j] = min({dp[i - 1][j - 1] + pxy , dp[i - 1][j] + pgap , dp[i][j - 1] + pgap }); } } } // Reconstructing the solution int l = n + m; // maximum possible length i = m; j = n; int xpos = l; int ypos = l; // Final answers for the respective strings int xans[l+1], yans[l+1]; while ( !(i == 0 || j == 0)) { if (x[i - 1] == y[j - 1]) { xans[xpos--] = ( int )x[i - 1]; yans[ypos--] = ( int )y[j - 1]; i--; j--; } else if (dp[i - 1][j - 1] + pxy == dp[i][j]) { xans[xpos--] = ( int )x[i - 1]; yans[ypos--] = ( int )y[j - 1]; i--; j--; } else if (dp[i - 1][j] + pgap == dp[i][j]) { xans[xpos--] = ( int )x[i - 1]; yans[ypos--] = ( int ) '_' ; i--; } else if (dp[i][j - 1] + pgap == dp[i][j]) { xans[xpos--] = ( int ) '_' ; yans[ypos--] = ( int )y[j - 1]; j--; } } while (xpos > 0) { if (i > 0) xans[xpos--] = ( int )x[--i]; else xans[xpos--] = ( int ) '_' ; } while (ypos > 0) { if (j > 0) yans[ypos--] = ( int )y[--j]; else yans[ypos--] = ( int ) '_' ; } // Since we have assumed the answer to be n+m long, // we need to remove the extra gaps in the starting // id represents the index from which the arrays // xans, yans are useful int id = 1; for (i = l; i >= 1; i--) { if (( char )yans[i] == '_' && ( char )xans[i] == '_' ) { id = i + 1; break ; } } // Printing the final answer cout << "Minimum Penalty in aligning the genes = " ; cout << dp[m][n] << "" ; cout << "The aligned genes are :" ; for (i = id; i <= l; i++) { cout<<( char )xans[i]; } cout << "" ; for (i = id; i <= l; i++) { cout << ( char )yans[i]; } return ; } // Driver code int main(){ // input strings string gene1 = "AGGGCT" ; string gene2 = "AGGCA" ; // initialising penalties of different types int misMatchPenalty = 3; int gapPenalty = 2; // calling the function to calculate the result getMinimumPenalty(gene1, gene2, misMatchPenalty, gapPenalty); return 0; } |
JAVA
// Java program to implement // sequence alignment problem. import java.io.*; import java.util.*; import java.lang.*; class GFG { // function to find out // the minimum penalty static void getMinimumPenalty(String x, String y, int pxy, int pgap) { int i, j; // initialising variables int m = x.length(); // length of gene1 int n = y.length(); // length of gene2 // table for storing optimal // substructure answers int dp[][] = new int [n + m + 1 ][n + m + 1 ]; for ( int [] x1 : dp) Arrays.fill(x1, 0 ); // initialising the table for (i = 0 ; i <= (n + m); i++) { dp[i][ 0 ] = i * pgap; dp[ 0 ][i] = i * pgap; } // calculating the // minimum penalty for (i = 1 ; i <= m; i++) { for (j = 1 ; j <= n; j++) { if (x.charAt(i - 1 ) == y.charAt(j - 1 )) { dp[i][j] = dp[i - 1 ][j - 1 ]; } else { dp[i][j] = Math.min(Math.min(dp[i - 1 ][j - 1 ] + pxy , dp[i - 1 ][j] + pgap) , dp[i][j - 1 ] + pgap ); } } } // Reconstructing the solution int l = n + m; // maximum possible length i = m; j = n; int xpos = l; int ypos = l; // Final answers for // the respective strings int xans[] = new int [l + 1 ]; int yans[] = new int [l + 1 ]; while ( !(i == 0 || j == 0 )) { if (x.charAt(i - 1 ) == y.charAt(j - 1 )) { xans[xpos--] = ( int )x.charAt(i - 1 ); yans[ypos--] = ( int )y.charAt(j - 1 ); i--; j--; } else if (dp[i - 1 ][j - 1 ] + pxy == dp[i][j]) { xans[xpos--] = ( int )x.charAt(i - 1 ); yans[ypos--] = ( int )y.charAt(j - 1 ); i--; j--; } else if (dp[i - 1 ][j] + pgap == dp[i][j]) { xans[xpos--] = ( int )x.charAt(i - 1 ); yans[ypos--] = ( int ) '_' ; i--; } else if (dp[i][j - 1 ] + pgap == dp[i][j]) { xans[xpos--] = ( int ) '_' ; yans[ypos--] = ( int )y.charAt(j - 1 ); j--; } } while (xpos > 0 ) { if (i > 0 ) xans[xpos--] = ( int )x.charAt(--i); else xans[xpos--] = ( int ) '_' ; } while (ypos > 0 ) { if (j > 0 ) yans[ypos--] = ( int )y.charAt(--j); else yans[ypos--] = ( int ) '_' ; } // Since we have assumed the // answer to be n+m long, // we need to remove the extra // gaps in the starting id // represents the index from // which the arrays xans, // yans are useful int id = 1 ; for (i = l; i >= 1 ; i--) { if (( char )yans[i] == '_' && ( char )xans[i] == '_' ) { id = i + 1 ; break ; } } // Printing the final answer System.out.print( "Minimum Penalty in " + "aligning the genes = " ); System.out.print(dp[m][n] + "" ); System.out.println( "The aligned genes are :" ); for (i = id; i <= l; i++) { System.out.print(( char )xans[i]); } System.out.print( "" ); for (i = id; i <= l; i++) { System.out.print(( char )yans[i]); } return ; } // Driver code public static void main(String[] args) { // input strings String gene1 = "AGGGCT" ; String gene2 = "AGGCA" ; // initialising penalties // of different types int misMatchPenalty = 3 ; int gapPenalty = 2 ; // calling the function to // calculate the result getMinimumPenalty(gene1, gene2, misMatchPenalty, gapPenalty); } } |
Python3
#!/bin/python3 """ Converted from C++ solution to Python3 Algorithm type / application: Bioinformatics Python Requirements: numpy """ import numpy as np def get_minimum_penalty(x: str , y: str , pxy: int , pgap: int ): """ Function to find out the minimum penalty :param x: pattern X :param y: pattern Y :param pxy: penalty of mis-matching the characters of X and Y :param pgap: penalty of a gap between pattern elements """ # initializing variables i = 0 j = 0 # pattern lengths m = len (x) n = len (y) # table for storing optimal substructure answers dp = np.zeros([m + 1 ,n + 1 ], dtype = int ) #int dp[m+1][n+1] = {0}; # initialising the table dp[ 0 :(m + 1 ), 0 ] = [ i * pgap for i in range (m + 1 )] dp[ 0 , 0 :(n + 1 )] = [ i * pgap for i in range (n + 1 )] # calculating the minimum penalty i = 1 while i < = m: j = 1 while j < = n: if x[i - 1 ] = = y[j - 1 ]: dp[i][j] = dp[i - 1 ][j - 1 ] else : dp[i][j] = min (dp[i - 1 ][j - 1 ] + pxy, dp[i - 1 ][j] + pgap, dp[i][j - 1 ] + pgap) j + = 1 i + = 1 # Reconstructing the solution l = n + m # maximum possible length i = m j = n xpos = l ypos = l # Final answers for the respective strings xans = np.zeros(l + 1 , dtype = int ) yans = np.zeros(l + 1 , dtype = int ) while not (i = = 0 or j = = 0 ): #print(f"i: {i}, j: {j}") if x[i - 1 ] = = y[j - 1 ]: xans[xpos] = ord (x[i - 1 ]) yans[ypos] = ord (y[j - 1 ]) xpos - = 1 ypos - = 1 i - = 1 j - = 1 elif (dp[i - 1 ][j - 1 ] + pxy) = = dp[i][j]: xans[xpos] = ord (x[i - 1 ]) yans[ypos] = ord (y[j - 1 ]) xpos - = 1 ypos - = 1 i - = 1 j - = 1 elif (dp[i - 1 ][j] + pgap) = = dp[i][j]: xans[xpos] = ord (x[i - 1 ]) yans[ypos] = ord ( '_' ) xpos - = 1 ypos - = 1 i - = 1 elif (dp[i][j - 1 ] + pgap) = = dp[i][j]: xans[xpos] = ord ( '_' ) yans[ypos] = ord (y[j - 1 ]) xpos - = 1 ypos - = 1 j - = 1 while xpos > 0 : if i > 0 : i - = 1 xans[xpos] = ord (x[i]) xpos - = 1 else : xans[xpos] = ord ( '_' ) xpos - = 1 while ypos > 0 : if j > 0 : j - = 1 yans[ypos] = ord (y[j]) ypos - = 1 else : yans[ypos] = ord ( '_' ) ypos - = 1 # Since we have assumed the answer to be n+m long, # we need to remove the extra gaps in the starting # id represents the index from which the arrays # xans, yans are useful id = 1 i = l while i > = 1 : if ( chr (yans[i]) = = '_' ) and chr (xans[i]) = = '_' : id = i + 1 break i - = 1 # Printing the final answer print (f "Minimum Penalty in aligning the genes = {dp[m][n]}" ) print ( "The aligned genes are:" ) # X i = id x_seq = "" while i < = l: x_seq + = chr (xans[i]) i + = 1 print (f "X seq: {x_seq}" ) # Y i = id y_seq = "" while i < = l: y_seq + = chr (yans[i]) i + = 1 print (f "Y seq: {y_seq}" ) def test_get_minimum_penalty(): """ Test the get_minimum_penalty function """ # input strings gene1 = "AGGGCT" gene2 = "AGGCA" # initialising penalties of different types mismatch_penalty = 3 gap_penalty = 2 # calling the function to calculate the result get_minimum_penalty(gene1, gene2, mismatch_penalty, gap_penalty) test_get_minimum_penalty() # This code is contributed by wilderchirstopher. |
C#
// C# program to implement sequence alignment // problem. using System; class GFG { // function to find out the minimum penalty public static void getMinimumPenalty( string x, string y, int pxy, int pgap) { int i, j; // initialising variables int m = x.Length; // length of gene1 int n = y.Length; // length of gene2 // table for storing optimal substructure answers int [,] dp = new int [n+m+1,n+m+1]; for ( int q = 0; q < n+m+1; q++) for ( int w = 0; w < n+m+1; w++) dp[q,w] = 0; // initialising the table for (i = 0; i <= (n+m); i++) { dp[i,0] = i * pgap; dp[0,i] = i * pgap; } // calculating the minimum penalty for (i = 1; i <= m; i++) { for (j = 1; j <= n; j++) { if (x[i - 1] == y[j - 1]) { dp[i,j] = dp[i - 1,j - 1]; } else { dp[i,j] = Math.Min(Math.Min(dp[i - 1,j - 1] + pxy , dp[i - 1,j] + pgap) , dp[i,j - 1] + pgap ); } } } // Reconstructing the solution int l = n + m; // maximum possible length i = m; j = n; int xpos = l; int ypos = l; // Final answers for the respective strings int [] xans = new int [l+1]; int [] yans = new int [l+1]; while ( !(i == 0 || j == 0)) { if (x[i - 1] == y[j - 1]) { xans[xpos--] = ( int )x[i - 1]; yans[ypos--] = ( int )y[j - 1]; i--; j--; } else if (dp[i - 1,j - 1] + pxy == dp[i,j]) { xans[xpos--] = ( int )x[i - 1]; yans[ypos--] = ( int )y[j - 1]; i--; j--; } else if (dp[i - 1,j] + pgap == dp[i,j]) { xans[xpos--] = ( int )x[i - 1]; yans[ypos--] = ( int ) '_' ; i--; } else if (dp[i,j - 1] + pgap == dp[i,j]) { xans[xpos--] = ( int ) '_' ; yans[ypos--] = ( int )y[j - 1]; j--; } } while (xpos > 0) { if (i > 0) xans[xpos--] = ( int )x[--i]; else xans[xpos--] = ( int ) '_' ; } while (ypos > 0) { if (j > 0) yans[ypos--] = ( int )y[--j]; else yans[ypos--] = ( int ) '_' ; } // Since we have assumed the answer to be n+m long, // we need to remove the extra gaps in the starting // id represents the index from which the arrays // xans, yans are useful int id = 1; for (i = l; i >= 1; i--) { if (( char )yans[i] == '_' && ( char )xans[i] == '_' ) { id = i + 1; break ; } } // Printing the final answer Console.Write( "Minimum Penalty in aligning the genes = " + dp[m,n] + "" ); Console.Write( "The aligned genes are :" ); for (i = id; i <= l; i++) { Console.Write(( char )xans[i]); } Console.Write( "" ); for (i = id; i <= l; i++) { Console.Write(( char )yans[i]); } return ; } // Driver code static void Main() { // input strings string gene1 = "AGGGCT" ; string gene2 = "AGGCA" ; // initialising penalties of different types int misMatchPenalty = 3; int gapPenalty = 2; // calling the function to calculate the result getMinimumPenalty(gene1, gene2, misMatchPenalty, gapPenalty); } //This code is contributed by DrRoot_ } |
PHP
<?php // PHP program to implement // sequence alignment problem. // function to find out // the minimum penalty function getMinimumPenalty( $x , $y , $pxy , $pgap ) { $i ; $j ; // initializing variables $m = strlen ( $x ); // length of gene1 $n = strlen ( $y ); // length of gene2 // table for storing optimal // substructure answers $dp [ $n + $m + 1][ $n + $m + 1] = array (0); // initialising the table for ( $i = 0; $i <= ( $n + $m ); $i ++) { $dp [ $i ][0] = $i * $pgap ; $dp [0][ $i ] = $i * $pgap ; } // calculating the // minimum penalty for ( $i = 1; $i <= $m ; $i ++) { for ( $j = 1; $j <= $n ; $j ++) { if ( $x [ $i - 1] == $y [ $j - 1]) { $dp [ $i ][ $j ] = $dp [ $i - 1][ $j - 1]; } else { $dp [ $i ][ $j ] = min( $dp [ $i - 1][ $j - 1] + $pxy , $dp [ $i - 1][ $j ] + $pgap , $dp [ $i ][ $j - 1] + $pgap ); } } } // Reconstructing the solution $l = $n + $m ; // maximum possible length $i = $m ; $j = $n ; $xpos = $l ; $ypos = $l ; // Final answers for // the respective strings // $xans[$l + 1]; $yans[$l + 1]; while ( !( $i == 0 || $j == 0)) { if ( $x [ $i - 1] == $y [ $j - 1]) { $xans [ $xpos --] = $x [ $i - 1]; $yans [ $ypos --] = $y [ $j - 1]; $i --; $j --; } else if ( $dp [ $i - 1][ $j - 1] + $pxy == $dp [ $i ][ $j ]) { $xans [ $xpos --] = $x [ $i - 1]; $yans [ $ypos --] = $y [ $j - 1]; $i --; $j --; } else if ( $dp [ $i - 1][ $j ] + $pgap == $dp [ $i ][ $j ]) { $xans [ $xpos --] = $x [ $i - 1]; $yans [ $ypos --] = '_' ; $i --; } else if ( $dp [ $i ][ $j - 1] + $pgap == $dp [ $i ][ $j ]) { $xans [ $xpos --] = '_' ; $yans [ $ypos --] = $y [ $j - 1]; $j --; } } while ( $xpos > 0) { if ( $i > 0) $xans [ $xpos --] = $x [-- $i ]; else $xans [ $xpos --] = '_' ; } while ( $ypos > 0) { if ( $j > 0) $yans [ $ypos --] = $y [-- $j ]; else $yans [ $ypos --] = '_' ; } // Since we have assumed the // answer to be n+m long, // we need to remove the extra // gaps in the starting // id represents the index // from which the arrays // xans, yans are useful $id = 1; for ( $i = $l ; $i >= 1; $i --) { if ( $yans [ $i ] == '_' && $xans [ $i ] == '_' ) { $id = $i + 1; break ; } } // Printing the final answer echo "Minimum Penalty in " . "aligning the genes = " ; echo $dp [ $m ][ $n ] . "" ; echo "The aligned genes are :" ; for ( $i = $id ; $i <= $l ; $i ++) { echo $xans [ $i ]; } echo "" ; for ( $i = $id ; $i <= $l ; $i ++) { echo $yans [ $i ]; } return ; } // Driver code // input strings $gene1 = "AGGGCT" ; $gene2 = "AGGCA" ; // initialising penalties // of different types $misMatchPenalty = 3; $gapPenalty = 2; // calling the function // to calculate the result getMinimumPenalty( $gene1 , $gene2 , $misMatchPenalty , $gapPenalty ); // This code is contributed by Abhinav96 ?> |
Minimum Penalty in aligning the genes = 5The aligned genes are :AGGGCTA_GGCA
时间复杂性: ![]() 空间复杂性:
空间复杂性: ![]()
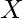

![关于”PostgreSQL错误:关系[表]不存在“问题的原因和解决方案-yiteyi-C++库](https://www.yiteyi.com/wp-content/themes/zibll/img/thumbnail.svg)



